Predicting Disease-related Genes Using Biomedical Literature Based on GloVe Word Embedding
*Senior Bioinformatician, Ebiogen PhD, Department of IT Convergence Engineering, Gachon University**Professor, Department of Computer Engineering, Gachon University
Abstract
Identifying disease-related genes is essential for understanding disease mechanisms and treating patients. Because wet-lab experiments are time-consuming and expensive, studies that use text mining have been increasing. In this paper, we propose a method to predict novel disease-related genes based on GloVe. Existing word embedding-based methods do not consider a characteristic of word embedding, so it is difficult to reproduce the experiment. However, the proposed method converges to a certain result through repeated experiments, it is possible to reproduce and more accurate. Furthermore, the proposed method can predict genes that are highly related to disease among genes that are not predicted by the existing methods and discover novel relationships based on daily produced bibliographic data.
초록
질병 관련 유전자를 식별하는 것은 질병 메커니즘을 이해하고 환자를 치료하는 데 필수적이다. 생물학적 실험은 시간과 비용이 많이 들기 때문에 텍스트 마이닝을 사용하는 연구의 수가 증가하고 있다. 본 연구는 GloVe기반으로 새로운 질병 관련 유전자를 예측하는 방법을 제안한다. 기존의 단어 임베딩 기반의 질병 연관 유전자 추론 방법들은 단어 임베딩의 특성을 고려하지 않았기 때문에 실험을 재현하기 매우 어렵다. 하지만 제안된 방법은 반복 실험을 통하여 일정한 결과에 수렴하기 때문에 실험 재현이 가능하며 기존 방법보다 정확하다. 마지막으로, 제안된 방법은 기존 방법에 따라 예측되지 않은 유전자 중 질병과 관련성이 높은 유전자를 예측할 수 있으며 계속해서 업데이트 되는 유전자-질병 연관을 통하여 새로운 관계를 발견할 수 있다.
Keywords:
text mining, word embedding, disease-gene association, computational biologyⅠ. Introduction
In the field of biology, considerable effort has been devoted to understanding the onset and treatment of diseases, where identifying disease-related genes is particularly important for understanding disease mechanisms and treating patients. For decades, researchers have proposed approaches for identifying disease-related genes. Because wet-lab experiments are time-consuming and expensive, performing research on all combinations of gene-disease associations is difficult [1]. Recently, studies using data mining-based computational methods have attempted to infer the relationships between genes and diseases and to predict novel associations. As the amount of available public data increases, computational methods can be used to acquire information more efficiently compared to traditional approaches [2]-[4].
Liu et al. proposed a method to construct a human brain-specific gene network by combining various genomic and proteomic data and to discover candidate genes sensitive to brain diseases [5]. They collected public microarray data, co-citation and protein-protein interaction (PPI) between brain genes for the human brain, and then built an integrated network based on the Bayesian statistical model. They identified crucial genes based on seed genes for specific diseases in the network and constructed a sub-network of those diseases using high-score interactions between genes. They predicted 46 susceptibility genes for Alzheimer’s disease and identified 23 genes as known genes related to this disease.
The goal of biomedical research is to discover novel and advanced biological information and apply this information to disease diagnosis, prevention, and treatment [6]. Text mining, which is the process of discovering interesting patterns or obtaining information from vast amounts of unstructured data, is an appropriate approach to accomplish such a task [7][8]. Text mining-based approaches can consistently acquire novel information considering that more than 100,000 sets of textual data are generated per day. In particular, various studies based on text mining have been proposed to discover gene-disease associations [9].
Kim et al. proposed a method to identify disease-related genes using Google search results and literature data [10]. From the literature, they collected sentences containing references to specific diseases and constructed a network by measuring the co-occurrence frequency of two genes. A node is a gene, and an edge is a co-occurrence frequency of two connecting genes in the network. They employed the Google search engine to measure the number of documents containing references to the two genes. They used the gene symbol as referenced in the literature as well as the full name of the gene derived from a Google search engine to measure the co-occurrence. They normalized the measured frequency and Google search results to a Z-score and then integrated and used two normalized scores to predict the candidate disease-related genes.
Word embedding (or word representation) is a natural language processing approach based on a neural network in which words are mapped to vectors of real numbers. Word embedding reflects the meaning and syntax of individual words in a low dimension [11]. Dimension reduction can help to overcome the sparsity of data, which is one of the limitations of traditional text mining approaches. Moreover, this approach is used to solve a variety of problems such as text classification and semantic analysis, as words in a vector space can be identified by word groups having similar meanings [12].
Koiwa et al. proposed a method of extracting disease-related genes from the literature using word2vec [13]. From the literature, they collected titles and abstracts about "schizophrenia", which is a psychiatric disorder from literature. They performed vectorization using a skip-gram, which is the one of the algorithms that execute word2vec. They then identified the top 10 words most similar to each known gene and predicted the gene symbols that emerged from the top words as candidate disease-related genes for schizophrenia. They predicted 62 novel disease-related genes using 461 known genes. They measured the classification performance using microarray data on schizophrenia to verify the validity of candidates.
Existing word embedding-based disease-related gene inference methods do not take into account the randomness of the initial embedding position, which is one of the characteristics of word embedding, so it is very difficult to reproduce the experiment. However, because the proposed method converges to a certain result through repeated experiments, it is possible to reproduce the experiment and it is more accurate than the existing method. The proposed method can predict genes that are highly related to disease among genes that are not predicted by the existing methods. Furthermore, the proposed method can continuously update gene-disease associations and discover novel relationships as considerable amount of bibliographic data are being produced daily.
Ⅱ. Materials and Methods
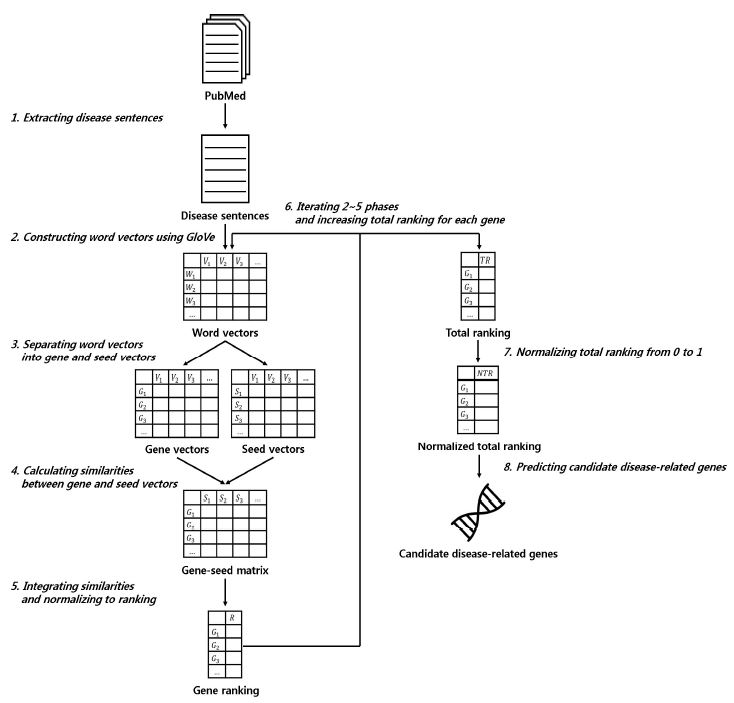
In this paper, we propose a method to predict novel disease-related genes using a vast amount of biological literature based on GloVe, one of the word embedding algorithms. GloVe integrates the characteristics of global matrix factorization (e.g., latent semantic analysis) and local context window methods (e.g., skip-gram) [14]. Global matrix factorization methods measure the word frequency in documents or sentences, where a row lists words and a column lists documents or sentences. These methods use statistical information but have difficulty in word analogy. Local context window methods predict target words by considering context words. These methods are strong for analogy but are limited by the fact that global frequency cannot be considered. GloVe is an algorithm that considers both the global frequency of words and the frequency within a context window. This method thus overcomes the limitations of existing methods. We collect sentences containing references to a specific disease from the abstracts of the literature and construct word vectors using GloVe. On the basis of known disease-related genes and the rest of the genes, we separate word vectors into gene vectors and seed vectors. We construct a gene-seed matrix by measuring the similarity between gene vectors and seed vectors. We calculate the sum of the similarities with the seed genes included in each gene in the gene-seed matrix and normalize the values of the genes to ranking. Since GloVe changes the result of the word vectors each time it is trained, these processes are repeated, and the total ranking is calculated and normalized to a value between 0 and 1. We predict candidate disease-related genes based on normalized total ranking. Fig. 1 shows the system overview of this paper.
2.1 Data description
PubMed is a database that provides literature data on various biomedical fields such as biomedicine, health, chemical science, and bioengineering [15]. Literature data are provided in XML format and include the title, authors, publication date, journal name, and abstract of each document. In this paper, we used only the abstracts from the literature. We collected 17,731,111 abstracts from 972 XML files provided by PubMed.
To identify a sentence containing reference to a disease, we needed the data regarding the name of the disease. We collected the official disease names and their synonyms from the Disease Ontology database [16][17]. Disease Ontology is a standardized ontology for human diseases that provides human disease terms, characteristics, and medical disease-associated vocabularies. We collected names and synonyms for specific diseases to verify the proposed method.
To extract gene vectors and seed vectors from word vectors, we collected gene symbols and known disease-related genes from the HUGO Gene Nomenclature Committee (HGNC) and Online Mendelian Inheritance in Man (OMIM) [18][19]. HGNC is a database that allows explicit scientific communication by providing a various information about genes. In this paper, we used 41,768 approved gene symbols out of 43,768 HGNC gene except 1,479 withdrawn symbols. Withdrawn gene symbols refer to a previously approved HGNC symbol for a gene that now has a different approved symbol.
OMIM is a comprehensive public database of human genes and genetic phenotypes. We defined known disease-related genes from OMIM as seed genes and used them to extract seed vectors from word vectors. Gene symbols constituting gene vectors and gene symbols constituting seed vectors did not overlap each other.
DisGeNet is a public database that contains information on disease-related genes and genetic mutations for human disease [20][21]. This database contains two categories of gene-disease associations, curated and text mining-based associations. Curated gene-disease associations are collected from UniProt, PsyGeNET, Orphanet, CGI, CTD, ClinGen, and the Genomic England PanelApp. We used curated gene-disease associations as classification label to validate the proposed method.
2.2 Extracting disease sentences
Sentences containing disease names were required for this paper. We collected abstracts from PubMed and separated them into sentence unit. For the purpose of separation, we used sentence boundary detection (SBD), which is a method of determining the beginning and end of a sentence in natural language processing [22][23]. Paragraphs were divided into sentences while considering the ambiguity of punctuation such as abbreviations and decimal points. Then, we extracted sentences containing references to specific disease names from the preprocessed sentences. We used the official disease name and its synonyms as given in the Disease Ontology database.
2.3 Constructing word vectors using GloVe
Using GloVe, we constructed word vectors based on the sentences collected for each disease. GloVe consists of three main phases: 1) Construct word co-occurrence matrix X. Xij is the frequency at which words i and j appear in the context. The window size of the context can be adjusted during frequency measurement. If the distance between the two words is long, lesser weight is given. 2) Define soft constraints for each word pair, which is shown in Equation 1.
| (1) |
Soft constraints are particularly useful to model and solve over-constrained and preference-based problems [24]. In Equation 1, Wi is a vector for the main word, Wj is a vector for the context word, bi and and bj are biases for main and context words, respectively. 3) Define the cost function, which is shown in Equation 2.
| (2) |
f is a weighting function applied to extremely frequent word pairs in Equation 2. f is weighted based on the condition of Equation 3.
| (3) |
In Equation 3, the default values for xmax and a are 100 and 3/4. GloVe generates two different word vectors, Wi and Wj. If X is a symmetric matrix, Wi is the same as Wj, and these matrices possess the properties whereby they differ only in terms of the results of random initialization. Consolidating the results of learning multiple instances of a network in a particular neural network can reduce overfitting and noise, thus improving results [14][25]. Therefore, we used the sum of Wi and Wj as the final vector in this paper.
GloVe requires various parameters for constructing word vectors. Window size refers to the range of words to be included in the context. For example, if the window size is 3, three words before and after the target word are considered. Vector size represents the dimension of the word vector. The larger the vector, the more accurate is the representation. However, the learning process requires considerable time. Minimum frequency refers to the minimum number of occurrences required for a word to be included in a word vector [26]. We constructed word vectors using "text2vec," which is an R package [27]. We derived the optimal parameters of GloVe from researches of Pennington et al. and Chiu et al [14][26].
2.4 Separating word vectors into gene and seed vectors
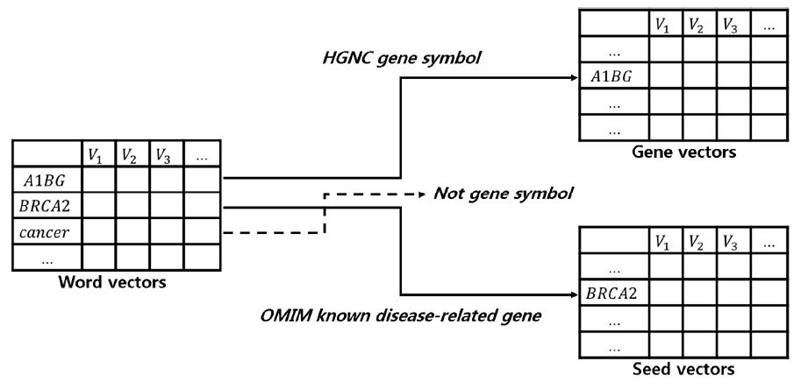
We separated word vectors that were built on sentences for a disease into gene and seed vectors. Gene vectors were composed of the same words as HGNC gene symbols among the words constituting word vectors. Seed vectors were constructed by the same words with known disease-related genes in OMIM for a disease among the words constituting word vectors. Gene symbols constituting gene vectors and seed vectors did not overlap each other. Fig. 2 shows the process of separating word vectors into gene and seed vectors for breast cancer.
In Fig. 2, V is a dimension in the word vectors. "A1BG" is the gene included in the HGNC gene symbol and "BRCA2" is the gene associated with breast cancer collected from OMIM. "BRCA2" is a gene included in HGNC, but it is not included in gene vectors because it is a gene constituting seed vectors. The word "cancer" is not considered in constructing either gene or seed vectors because it does not appear in the HGNC and OMIM.
2.5 Calculating similarities between gene and seed vectors
We measured similarity between gene and seed for a disease using gene vectors and seed vectors. It is based on the precondition that if a gene is similar to a seed, the gene is likely to be a candidate disease-related gene. We calculated the similarity by measuring cosine similarity. Equation 4 is the formula for calculating the similarity between gene and seed using their vectors.
| (4) |
In Equation 4, G is the gene, S is the seed, and v is the number of dimensions in word vectors. We measured the similarity of all gene pairs in gene vectors and seed vectors using Equation 4 and constructed a gene-seed matrix.
2.6 Integrating similarities and normalizing to ranking
After the gene-seed matrix was constructed for a disease, we measured the average of all similarities for each gene and ranked.
Since the distribution of measured similarities differs for each seed, we normalized the mean of similarities to ranking. We assigned higher value of ranking to gene with higher mean similarity to seeds, and lower value of ranking to gene with lower mean similarity. In the case of 10 genes, for example, 10 is assigned to the gene having the highest mean similarity and 1 is assigned to the gene having the lowest mean similarity.
2.7 Increasing and normalizing total ranking for each gene
Because word vectors are constructed differently each time when the model is trained, the similarities are calculated differently. It causes bias in the overall experimental results. In order to overcome this limitation, we repeated the process from constructing word vectors to integrating similarities and normalizing to ranking. The more iterations, the greater the accuracy of the results, but it increases the time required for the experiment. In this study, we repeated the preceding phases 30 times. We measured the total ranking for the genes calculated over 30 iterations and normalized the total ranking to a value between 0 and 1.
2.8 Predicting candidate disease-related genes
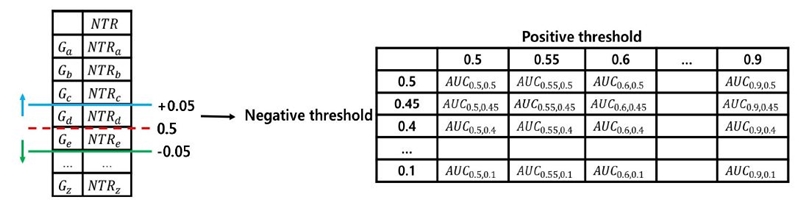
We constructed a classifier to predict disease-related genes using normalized total ranking. First, we sorted the normalized total ranking in descending order. We assigned "True" class label to gene with values greater than 0.5 and "False" to genes with values less than 0.5. Next, we measured the classification performance by increasing and decreasing the value by 0.05 starting at 0.5. We named the "True" class threshold as the positive threshold and "False" as negative threshold. Of all the combinations of positive and negative thresholds, we identified the thresholds with the highest AUC and used this case as a classifier to predict disease-related genes. Because the combination of thresholds with an AUC of 1 can occur when there is a small number of data while measuring classification performance, the combination with the highest AUC with a less than 1 is used as the thresholds of the classifier. Fig. 3 shows the process of identifying the optimal thresholds for constructing the classifier.
In Fig. 3, Ga,…,Gz and NTRa,…,NTRz are genes and their values sorted in descending order of normalized total ranking. The red dotted line is the portion where the normalized total ranking is 0.5. The blue line is positive threshold and the green line is negative threshold. AUCp,n is an AUC value for positive threshold p and negative threshold n. AUC was measured as the threshold was adjusted, and the combination with the highest AUC value other than 1 was used as the positive and negative thresholds for classifying disease-related genes. While adjusting the positive threshold and the negative threshold by 0.05 each, AUC was measured for all combinations, and the optimal threshold was specified for the highest AUC.
Ⅲ. Results and discussion
3.1 Experimental data
To evaluate the proposed method and predict novel disease-related genes, we selected cancers that could be applied to the proposed method out of 36 cancers with high frequency in 185 countries [28].
We chose diseases under the condition that 1) disease-related genes should be provided in OMIM and 2) more than five seeds should appear in word vectors constructed from disease sentences. In other words, we only selected disease for which there is a sufficient number of seeds. We collected sentences from PubMed that include lung cancer, breast cancer, prostate cancer, colon cancer (and colorectal cancer), stomach cancer, liver cancer, and bladder cancer among 36 cancers. Table 1 shows the number of sentences containing each disease.
While constructing the word vectors, we applied the optimal parameters in Table 2 to model training process of GloVe.
3.2 Optimal positive and negative threshold
We tuned the positive and negative threshold for each disease to identify optimal thresholds and construct a classifier with highest AUC score. Table 3 shows the optimal threshold of the positive and negative, and AUC for the combination of the thresholds for each disease.
For each disease, we assigned "True" label to genes with a value greater than the positive threshold and "False" label to genes with a value less than the negative threshold in Table 3.
3.3 Comparison with other methods
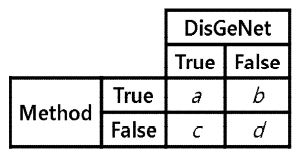
We compared the classification performance of the proposed method with those of other disease-related gene inference methods, such as DTMiner and RENET [29][30]. DTMiner is an approach to assessing how closely a gene-disease pair is related from a sentence in which genes and diseases appear simultaneously based on NER (Named entity recognition). RENET is a gene-disease inference method that considers classification at both sentence and document-level. RENET infers true associations among candidate gene-disease associations through the NER and RE (Relation extraction) steps. We constructed a confusion matrix based on the predicted true/false associations for each disease and curated gene-disease associations collected from DisGeNet, and measured the precision, recall and F-score for each method and compared the classification performance. Fig. 4 shows the confusion matrix, and Equation 5, 6, and 7 show the precision, recall, and F-score, respectively.
| (5) |
| (6) |
| (7) |
We confirmed that the proposed method has higher classification performance than the existing methods for seven diseases except liver cancer through Table 4. Liver cancer has much fewer disease sentences than other diseases as mentioned in Table 1, so we confirmed that liver cancer has low performance because the number of sentences is not enough to construct word vectors.
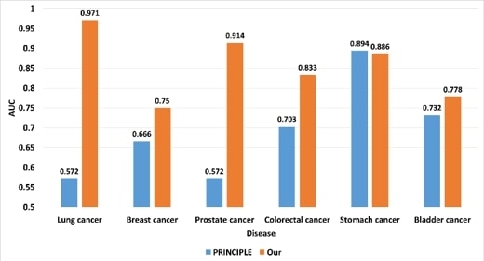
We compared the classification performance of the proposed method and the network-based disease association inference method. PRINCIPLE prioritizes disease-related genes based on closeness in a PPI network for disease query, and this method is executed as a plug-in to the Cytoscape [31]-[33].
We compared the classification performance for six of the eight diseases that resulted in PRINCIPLE. In PRINCIPLE, we collected the top 100 genes and their scores most relevant to each disease, and we assigned DisGeNet curated gene-disease associations as class labels. Fig. 5 shows the AUC comparison between PRINCIPLE and the proposed method.
We confirmed that the classification accuracy of the proposed method for five of the six disease is high from Fig. 5.
PRINCIPLE requires disease-disease similarity and known gene-disease associations to construct the network. The proposed method has the advantage the no additional information about the disease or gene is needed because only known disease-related genes are considered to infer novel disease-related genes.
3.4 The number of predicted disease-related genes
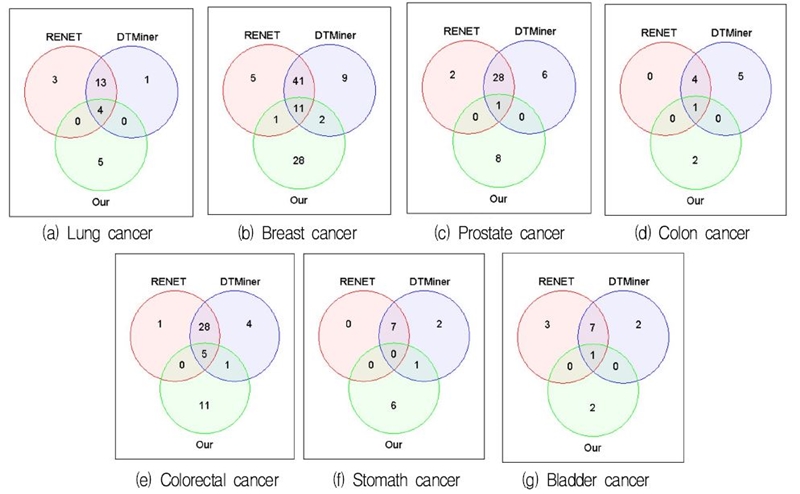
We compared the number of disease-related genes for each disease predicted by the proposed method with RENET and DTMiner. We compared the number of genes inferred in each method for seven diseases except liver cancer, which had the lowest classification performance of the proposed method. Fig. 6 shows the Venn diagram for the predicted number of genes for seven diseases.
In Fig. 6, the red area represents the number of predicted disease-related genes by RENET, the blue represents DTMiner, and the green represents the proposed method. Because RENET and DTMiner are sentence-level approaches, we confirmed that most of the disease-related genes predicted by the two methods overlap. The proposed method predicts fewer number of genes in total, but more genes in its own.
3.5 Literature-based evaluation of novel disease-related genes
We made a literature-based evaluation of novel disease-related genes inferred for breast cancer. We predicted 11 novel genes for breast cancer. Table 5 shows the novel disease-related genes, descriptions, and PMIDs for breast cancer discovered by the proposed method.
Ⅳ. Conclusion
We proposed a novel method for predicting disease-related genes from a vast amount of biomedical literature based on GloVe. We constructed word vectors using sentences containing disease and constructed a classifier based on the similarity between gene vectors and seed vectors.
We evaluated the proposed method with various diseases and derived candidate disease-related genes for breast cancer among these diseases.
We confirmed that the proposed method outperforms existing literature-based disease-related gene inference methods. Existing word embedding-based disease-related gene inference methods do not take into account the randomness of the initial embedding position, which is one of the characteristics of word embedding, so it is very difficult to reproduce the experiment. However, because the proposed method converges to a certain result through repeated experiments, it is possible to reproduce the experiment and it is more accurate than the existing method. The proposed method can predict genes that are highly related to disease among genes that are not predicted by the existing methods. Furthermore, the proposed method can continuously update gene-disease associations and discover novel relationships as considerable amount of bibliographic data are being produced daily.
However, the proposed method has the limitation that the experiment can be performed only for diseases with a sufficient number of seeds. Eight out of 36 cancers could be used for this paper. In future research, we will apply an approach to derive novel disease-related genes, regardless of the number of seed for the disease.
Acknowledgments
This research was supported by a grant from the National Research Foundation of Korea (NRF), funded by the Government of Korea (Ministry of Science, ICT and Future Planning) (NRF-2018R1A2B6006223).
References
-
J. Zhou and F. Bo-quan, "The research on gene-disease association based on text-mining of PubMed", BMC bioinformatics, Vol. 19, No. 1, pp. 37, Dec. 2018.
[https://doi.org/10.1186/s12859-018-2048-y]

-
S. W. Shin, Y. E. Shin, G. U. Jang, and Y. M. Yoon, "Co-occurrence Based Drug-disease Relationship Inference with Genes as Mediators", Journal of KIIT, Vol. 16, No. 11, pp. 1-9, Nov. 2018.
[https://doi.org/10.14801/jkiit.2018.16.11.1]

-
G. U. Jang, Y. H. Hwang, M. Oh, T. K. Lee, and Y. M. "Novel Drug Similarity Measuring Method based on Text Mining", Journal of KIIT, Vol. 14, No. 7, pp. 127-137, Jul. 2016.
[https://doi.org/10.14801/jkiit.2016.14.7.127]

-
Y. Keon, H. Kim, J.Y. Choi, D. Kim , S.Y. Kim, and S. Kim, "Call Center Call Count Prediction Model by Machine Learning", Journal of Advanced Information Technology and Convergence, Vol. 8, No. 1, pp. 31-42, Jul. 2018.
[https://doi.org/10.14801/JAITC.2018.8.1.31]

-
B. Liu, T. Jiang, S. Ma, H. Zhao, J. Li, X. Jiang, and J. Zhang, "Exploring candidate genes for human brain diseases from a brain-specific gene network", Biochemical and biophysical research communications, Vol. 349, No. 4, pp. 1308-1314, Nov. 2006.
[https://doi.org/10.1016/j.bbrc.2006.08.168]

-
A. M. Cohen and W. R. Hersh, "A survey of current work in biomedical text mining", Briefings in bioinformatics, Vol. 6, No. 1, pp. 57-71, Mar. 2005.
[https://doi.org/10.1093/bib/6.1.57]

- A. H. Tan, "Text mining: The state of the art and the challenges", In Proceedings of the PAKDD 1999 Workshop on Knowledge Disocovery from Advanced Databases, Vol. 8, pp. 65-70, Apr. 1999.
- J.U. Heu, "Korean Language Clustering Using Word2vec.", The Journal of Institute of Internet, Broadcasting and Communication, Vol. 18, No. 5 pp. 25-30, Oct. 2018.
-
T. Ono and S. Kuhara, "A novel method for gathering and prioritizing disease candidate genes based on construction of a set of disease-related MeSH® terms", BMC bioinformatics, Vol. 15, No. 1, pp. 179, Dec. 2014.
[https://doi.org/10.1186/1471-2105-15-179]

-
J. Kim, H. Kim, Y. Yoon, and S. Park, "LGscore: a method to identify disease-related genes using biological literature and Google data", Journal of biomedical informatics, Vol. 54, pp. 270-282, Apr. 2015.
[https://doi.org/10.1016/j.jbi.2015.01.003]

-
O. Melamud, J. Goldberger, and I. Dagan, "Context2vec: Learning generic context embedding with bidirectional lstm", In Proceedings of The 20th SIGNLL Conference on Computational Natural Language Learning, Berlin, Germany, pp. 51-61, Aug. 2016.
[https://doi.org/10.18653/v1/K16-1006]

-
J. Lilleberg, Y. Zhu, and Y. Zhang, "Support vector machines and word2vec for text classification with semantic features", In 2015 IEEE 14th International Conference on Cognitive Informatics & Cognitive Computing (ICCI* CC), Beijing, China, pp. 136-140, Jul. 2015.
[https://doi.org/10.1109/ICCI-CC.2015.7259377]

-
T. Koiwa and H. Ohwada, "Extraction of disease-related genes from PubMed paper using word2vec", In Proceedings of the 8th International Conference on Computational Systems-Biology and Bioinformatics, Nha Trang City Viet Nam, pp. 46-49, Dec. 2017.
[https://doi.org/10.1145/3156346.3156355]

-
J. Pennington, R. Socher, and C. D. Manning, "Glove: Global vectors for word representation", In Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP), Doha, Qatar, pp. 1532-1543, Oct. 2014.
[https://doi.org/10.3115/v1/D14-1162]

- K. Canese and S. Weis, "PubMed: the bibliographic database", The NCBI Handbook, pp. 2-1, Mar. 2013.
-
L. M. Schriml, C. Arze, S. Nadendla, Y. W. W. Chang, M. Mazaitis, V. Felix, G. Feng, and W. A. Kibbe, "Disease Ontology: a backbone for disease semantic integration", Nucleic acids research, Vol. 40, No. D1, pp. D940-D946, Jan. 2012.
[https://doi.org/10.1093/nar/gkr972]

-
W. A. Kibbe, C. Arze, V. Felix, E. Mitraka, E. Bolton, G. Fu, C. J. Mungall, J. X. Binder, J. Malone, D. Vasant, and H. Parkinson, "Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data", Nucleic acids research, Vol. 43, No. D1, pp. D1071-D1078, Jan. 2015.
[https://doi.org/10.1093/nar/gku1011]

-
S. Povey, R. Lovering, E. Bruford, M. Wright, M. Lush, and H. Wain, "The HUGO gene nomenclature committee (HGNC)", Human genetics, Vol. 109, No. 6, pp. 678-680, Dec. 2001.
[https://doi.org/10.1007/s00439-001-0615-0]

-
A. Hamosh, A. F. Scott, J. S. Amberger, C. A. Bocchini, and V. A. McKusick, "Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders", Nucleic acids research, Vol. 33, pp. D514-D517, Jan. 2005.
[https://doi.org/10.1093/nar/gki033]

-
J. Piñero, J. M. Ramírez-Anguita, J. Saüch-Pitarch, F. Ronzano, E. Centeno, F. Sanz, and L. I. Furlong, "The DisGeNET knowledge platform for disease genomics: 2019 update", Nucleic acids research, Vol. 48, No. D1, pp. D845-D855, Jan. 2020.
[https://doi.org/10.1093/nar/gkz1021]

-
J. Piñero, À. Bravo, N. Queralt-Rosinach, A. Gutiérrez-Sacristán, J. Deu-Pons, E. Centeno, J. García-García, F. Sanz, and L. I. Furlong, "DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants", Nucleic acids research, pp. gkw943, Oct. 2016.
[https://doi.org/10.1093/nar/gkw943]

-
D. Gillick, "Sentence boundary detection and the problem with the US", In Proceedings of Human Language Technologies: The 2009 Annual Conference of the North American Chapter of the Association for Computational Linguistics, Boulder, Colorado, pp. 241-244, Jun. 2009.
[https://doi.org/10.3115/1620853.1620920]

- Sentence Boundary Detection (SBD), https://github.com/lukeorland/splitta, . [accessed: Sep. 09, 2019]
-
W. J. van Hoeve and I. Katriel, "Global constraints", In Foundations of Artificial Intelligence, Vol. 2, pp. 169-208, Jan. 2006.
[https://doi.org/10.1016/S1574-6526(06)80010-6]

- D. Ciresan, A. Giusti, L. M. Gambardella, and J. Schmidhuber, "Deep neural networks segment neuronal membranes in electron microscopy images", In Advances in neural information processing systems, pp. 2843-2851, 2012.
-
B. Chiu, G. Crichton, A. Korhonen, and S. Pyysalo, "How to train good word embeddings for biomedical NLP", In Proceedings of the 15th workshop on biomedical natural language processing, pp. 166-174, Aug. 2016.
[https://doi.org/10.18653/v1/W16-2922]

- text2vec.org
-
F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre, and A. Jemal, "Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries", CA: a cancer journal for clinicians, Vol. 68, No. 6, pp. 394-424, Sep. 2018.
[https://doi.org/10.3322/caac.21492]

- D. Xu, M. Zhang, Y. Xie, F. Wang, M. Chen, K. Q. Zhu, and J. Wei, "DTMiner: identification of potential disease targets through biomedical literature mining", Bioinformatics, Vol. 32, No. 23, pp. 3619-3626, Dec. 2016.
-
Y. Wu, R. Luo, H. C. Leung, H. F. Ting, and T. W. Lam, "Renet: A deep learning approach for extracting gene-disease associations from literature", In International Conference on Research in Computational Molecular Biology, Padua, Italy, pp. 272-284, May 2019.
[https://doi.org/10.1007/978-3-030-17083-7_17]

-
O. Vanunu, O. Magger, E. Ruppin, T. Shlomi, and R. Sharan, "Associating genes and protein complexes with disease via network propagation", PLoS computational biology, Vol. 6, No. 1, pp. 1-9, Jan. 2010.
[https://doi.org/10.1371/journal.pcbi.1000641]

-
A. Gottlieb, O. Magger, I. Berman, E. Ruppin, and R. Sharan, "PRINCIPLE: a tool for associating genes with diseases via network propagation", Bioinformatics, Vol. 27, No. 23, pp. 3325-3326, Dec. 2011.
[https://doi.org/10.1093/bioinformatics/btr584]

-
P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski, and T. Ideker, "Cytoscape: a software environment for integrated models of biomolecular interaction networks", Genome research, Vol. 13, No. 11, pp. 2498-2504, Nov. 2003.
[https://doi.org/10.1101/gr.1239303]


2016 : BS degree in Department of Computer Engineering
2016 ~ 2020 : PhD degree in Department of IT Convergence Engineering, Gachon University.
2020. 01 ~ present : Senior Bioinformatician, Ebiogen
Research interests : text mining, systems, word embedding, computational biology

1981 : BS degree from Seoul National University
2008 : PhD degree in Department of Computer Science, Yonsei University.
1995 ~ present : Professor, Department of Computer Engineering, Gachon University
Research interests : database, data science, data mining, and bioinformatics 1