A MA-plot-based Feature Selection by MRMR in SVM-RFE in RNA-Sequencing Data
Abstract
It is extremely lacking and urgently required that the method of constructing the Gene Regulatory Network (GRN) from RNA-Sequencing data (RNA-Seq) because of Big-Data and GRN in Big-Data has obtained substantial observation as the interactions among relevant featured genes and their regulations. We propose newly the computational comparative feature patterns selection method by implementing a minimum-redundancy maximum-relevancy (MRMR) filter the support vector machine-recursive feature elimination (SVM-RFE) with Intensity-dependent normalization (DEGSEQ) as a preprocessor for emphasizing equal preciseness in RNA-seq in Big-Data. We found out the proposed algorithm might be more scalable and convenient because of all libraries in R package and be more improved in terms of the time consuming in Big-Data and minimum-redundancy maximum-relevancy of a set of feature patterns at the same time.
초록
유전자 규정 네트워크 (GRN)에 RNA-시퀀싱 데이터를 활용할 때, 해당 유전자와 환경과의 상호 작용에 의해서 생기는 형질들 중에서 연관성이 높은 유전자로 GRN을 구성하는 것은 상당히 어려운 일이다. 본 연구에서는 Big-Data의 RNA-시퀀싱 자료들로, 지지 벡터 머신 회귀 특징 추출(SVM-RFE) 에 근거하여, 연관성이 높은 유전자(maximum-relevancy)는 추출하고, 연관성이 낮은 유전자(minimum-redundancy)는 제거하는 MRMR 필터 방법을 집중도 의존 정규화(intensity-dependent normalization, DEGSEQ)에 기반 하여 데이터의 정밀성을 높여, 소수 연관성 높은 유전자만 판별해 내는 방법을 사용한다. 제안한 방법은 R 언어 패키지를 사용하여 편리함과 동시에, 다른 기존의 방법을 비교하였을 때, Big-Data의 시간 활용도를 높이면서, 동시에 높은 연관성 있는 유전자만을 잘 추출해 냄을 확인하였다.
Keywords:
SVM-RFE, MA-plot-based methods, RNA-Seq, MRMR, DEGⅠ. Introduction
Determining differential-expressed genes (DEGs) and building GRN provide improvement in both quality and quantity for reviewing the intricately obscure feature patterns of gene interaction [1][2]. The fundamentally required GRN have been to maximize distinguishable and minimize relevant feature patterns in genes to provide a primary element for molecular mechanisms through the inferential causality [1][2]. Although RNA-seq generates a huge amount of data, few computational methods can exploit the huge of Big-Data, such as NCBI-GEO [3][4]. It is extremely lacking and urgently required that the method of constructing of the GRN from RNA-Seq. However, it has been significantly challengeable to select subset genes of MRMR [5] because of the high dimensional and a low sample biological data [6]. That means the size of subset genes is extremely larger than those of samples. We call it that “Curse of Dimensionality” [6][7]. Guyon. et. al. proposed SVM-RFE [7] to eliminate irrelevant featured pattern genes on the weight vectors and classify featured samples with SVM. However, there are some flaws in SVM-RFE for determining differential-expressed genes (DEGs) from RNA-seq [1][2]. Therefore, we suggest a novel gene selection, which can exploit MRMR in SVM-RFE for mutual information between genes and MA-plot-based (Intensity-dependent normalization based on standard-deviations) in RNA-Seq [2] for emphasizing even preciseness and accuracy in Big-Data. We can exploit DEGSEQ, a free R package for the MA-plot-based.
Some previous researches developed the alternative algorithms based on SVM-RFE, for overcoming the consumption of the huge training time because of only one gene per iteration and over-fitting and improving the accuracy with narrowing down the potential set of cancerous genes [6]-[10]. For example, in Liu. et. al.[6], for overcoming over-fit feature patterns, they suggest the deep neural networks. However, in terms of small samples, they use multiple randomly drops neurons and features. Therefore, there are still rooms for the development based on mutual information for higher accuracy. Therefore, in the proposed algorithm, MRMR [5] can be exploited in SVM-RFE, which also uses the MA-plot-based (DEGSEQ) to improve the accuracy with decreasing the number of potentially relevant genes. We found out the proposed algorithm might be more scalable and convenient because of all libraries in R for computational resources. We can compare evaluations of the proposed algorithm with the SVM-RFE [7] on public gene expressions [3][4]. The comparison was outperformed than the previous one because the developed algorithms, Tang. et. al [8] might not consider the Big-Data because of SVM-RFE with Two-Stages which needs considerably huge computational resources. We can find that our algorithm might be performing with smaller computational resources with quite significant accuracy. The rest of the paper is organized as follows. The materials and methods are in Section 2. Section 3 describes the proposed algorithm. Section 4 gives the experimental results and comparison. Finally, we conclude our work in Section 5.
Ⅱ. Materials and Methods
2.1 Data
For comparison of the proposed algorithm with the previous Golub. et. al, we downloaded AML/ALL leukemia [3]. We analyzed AML/ALL by R-package “golubEsets” and “e1071” for SVM-RFE and DEGSEQ.
2.2 SVM-RFE algorithm
Guyon et al. proposes a feature pattern selection, SVM-RFE [7]. When facing highly dimensional and the low size of samples, classification or prediction problem suffers from over-fitting and high-variance gradients [8]. However, some machine learning algorithms likewise SVM-RFE can make good results on the low size of samples with low-variance gradients. SVM-RFE removes irrelevant the gene that is the smallest ranking criteria from the gene set [7]. The criteria for the score of gene ranking is used as the measurement of the determinant of featured genes. The weight vector w of the SVM defines the gene ranking score,
where w is calculated as.
| (1) |
where xi is the gene expression vector of a sample i in the training set, yi is the class label of i, yi ∊[-1, 1] and ai is the “Lagrangian Multiplier”. With a non-zero weight of vectors, ai are support vectors [7].
2.3 MA-plot-based
The MA-plot-based is a statistical analysis for detecting and visualizing the intensity-dependent normalization ratio of micro-array data [1]-[4]. The assayed micro-array data, every single one of them has been enormously noised because of the individual experiments. The intensity normalization uses Minus vs Add plot (M = log2C1− log2C2 and A = (log2C1+ log2C2)/2, where C1 and C2 is for exploration of the locally weighted linear regression) [1]. DEGSEQ of R packages[1] is the plot method for the above-mentioned purpose. DEGSEQ is color-coding the expressed genes based on 3 categories of standard-deviations.
2.4 MRMR algorithm
In MRMR, if an expressed feature pattern is in random or uniform distribution within different classes, its mutual information with these classes is zero [5]. Therefore, we can exploit the information as a determination of relevancy of expressed feature patterns. For feature patterns, mutual information, I of x and y is determined by joint probabilistic distribution, p(x,y) and respective marginal probabilities, p(x) and p(y) [5][9]]:
| (2) |
For feature patterns, we exploit I for determining the similarity among feature patterns [5]. The MRMR is for targeting the feature patterns having mutual maximal dissimilar [5]. Let S define as the featured subset. The minimum redundancy is [5][9]
| (3) |
where I(i,j) as I(h,gi) for simplicity [5][9]. To determine for discrimination of feature patterns for differential-expressed of different targets, we exploit I(h,gi) between targets, = {h1, h2, . . . , hK}, where h is classified target patterns and determines the gene gi' relevancy for the classified group. The maximum relevance is to maximize the relevancy of the all-featured patterns in S in total [5][9]:
| (4) |
I(h,gi) is I(h,I). The MRMR is indicated by two simplest combination criteria [5][9], (a)Mutual Information Different criterion(MID): max(VI−WI) or, (b)Mutual Information Quotient criterion(MIQ): max(VI/WI).
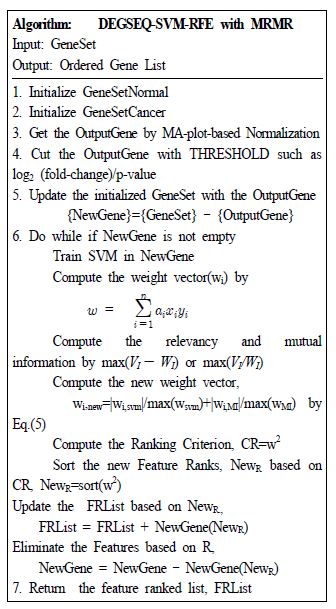
Ⅲ. The proposed Algorithm
In the proposed algorithm of Fig. 1, there will be the identified topmost ranking gene lists by the MA-plot-based in SVM-RFE with MRMR. For this purpose, MRMR [5] can be used as ranking criteria in SVM-RFE [7]. The SVM-RFE [7] trains the algorithm in every iteration for eliminating a feature patterns per time with different feature patterns. The SVM-RFE [7] has some flaws, that needs considerable time for their machine learning for training because of removing the redundant feature pattern per iteration. The previous researches implicitly assumes that the final-ranked feature patterns might be better if SVM-RFE eliminate a feature pattern per iteration.
Tang. et. al. [8] enhances SVM-RFE with Two-Stages. Therefore, the research [8] needs huge computational resources more than the previous one and hyper-parameters with system designers’ interactions. However, some previous researches assure that the assumption might not be always accurate when it emulated with AML/ALL [3]. So we exploit the MRMR in the SVM-RFE for different criterion as a filter-out for better computational performance. For further enhanced filter-out, we can exploit the MA-plot-based by MA-plot-based before the SVM-RFE with MRMR instead of Two-Stages. The proposed SVM-RFE with MRMR based on MA-plot-based for aiming for the better qualified training in terms of machine learning. In the MA-plot-based by MRMR in SVM-RFE, the new criterion is,
| (5) |
Ⅳ. The performance comparisons
Recently, researches emphasize that a smaller perfect feature patterns as a target are selected for better accurate validation [6]-[10]. The smaller selected, the better claimed. However, the smaller feature patterns might not justify the feature target selection methods outstanding compared to other ones in terms of extracting highly correlated feature patters because of “Curse of Dimension”[6]. Moreover, smaller feature patterns might not be extractable within a comparative computational performance. Therefore, we try to extract the distinguishable feature patterns that describe the complicated gene regulation in AML/ALL with regard to the computational strengthen.
Fig. 2 shows that comparition of the proposed algorithm with the previous one [7] based on the SVM-RFE with Two-Stages [8]. We describe how many distinguishable feature patterns are ranked in almost same levels. The all-feature patterns selected from [8] might not be in our result. However, “gene X95735” (Home sapience Zyxin) is ranked in the same place, the 3rd. And there are topmost genes in the highest place, for example, “gene M96326” (Azurocidin).
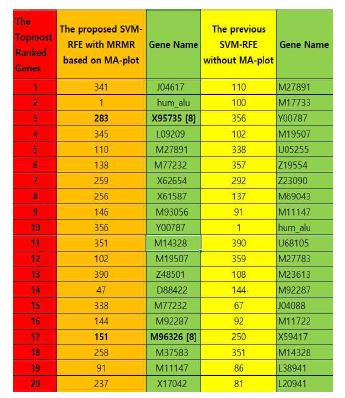
Comparition of the proposed algorithm with the previous SVM-RFE[4] based on SVM-RFE with Two-Stages[8]
Moreover, our recent research, C. Kim[10] is regard to the SVM-RFE enhanced with Time-Difference(T-D) Reinforcement Learning(RL). The results of the research [10] are regard to “how many feature patterns in the topmost rank list”. So those have quantitative worth. However, in our result in Fig. 2 are following up the question, “how many feature patterns in the same places in the rank list”. Therefore, we can find out that the proposed algorithm can improve the computational performance by enhancing the previous SVM-RFE [7] for a better qualified machine learning in terms of Big-Data. Normally, it was considered that the feature patterns might not be accurate if samples with high dimensionality are being used. However, Fig. 2 shows that the feature patterns of the proposed algorithm are comparable with the results of Golub. et. al [3] and shows as a quite computational comparative.
Ⅴ. Conclusion
We suggest a novel gene selection algorithm, which can exploit MRMR for maximum-relevancy and minimum-redundancy in SVM-RFE right after the MA-plot-based, which uses Minus vs Add plot by DEGSEQ in R package in RNA-Seq. The proposed algorithm can improve the accuracy with decreasing the number of potential irrelevant subset genes, respecting the computational comparative. For more accuracy, MRMR can be used as ranking criteria in SVM-RFE. The proposed algorithm can be convenient because of all libraries in R package. The comparison outperformed than the previous one in terms of Big-Data because some distinguishable feature patterns are in the same rank with a better qualified machine learning by enhancing criteria, MRMR based on MA-plot-based. For future works, we focus on the off-policy reinforcement learning (RL) with experience replay to learn to how to classify the real-time data either ‘normal’ or ‘abnormal’.
References
-
H. Bolouri, "Modeling genomic regulatory networks with big data", Trends in Genetics, 30(5), p182, May), (2014.
[https://doi.org/10.1016/j.tig.2014.02.005]

- Y. H. Yang, S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed, "Normalization for cDNA microarray data", Nucleic Acids Res, 30(4), p(e)15, Feb), (2002.
-
T. R. Golub, D. K. Slonim, P. Tamayo, C. Huard, M. Gaasenbeek, J. P. Mesirov, H. Coller, M. L. Loh, J. R. Downing, M. A. Caligiuri, C. D. Bloomfield, and E. S. Lander, "Molecular classification of cancer: class discovery and class prediction by gene expression monitoring", Science, 286(5439), p531-537, Oct), (1999.
[https://doi.org/10.1126/science.286.5439.531]

-
S. K Kim, S. Y Kim, J. H Kim, S. A Roh, D. H Cho, Y. S Kim, and J. C Kim, "A nineteen gene-based risk score classifier predicts prognosis of colorectal cancer patients", Molecular Oncology, 8(8), p653-1666, Dec), (2014.
[https://doi.org/10.1016/j.molonc.2014.06.016]

-
C. Ding, and H. Peng, "Minimum Redundancy Feature Selection from Microarray Gene Expression Data", J. Bioinfo. Compu. Bio., 3(2), p185-205, Apr), (2005.
[https://doi.org/10.1142/s0219720005001004]

- B. Liu, Y. Wei, Y. Zhang, and Q. Yang, "Deep Neural Networks for High Dimension, Low Sample Size Data", IJCAI-17, p2287-2293, Aug), (2017.
- I. Guyon, J. Weston, S. Barnhill, V. Vapnik, "Gene selection for cancer classification using support vector machine", Mach. Learn, 46, p389-422, Jan), (2002.
-
Y. Tang, Y. Q. Zhang, and Z. Huang, "Development of Two-Stage SVM-RFE Gene Selection Strategy for Microarray Expression Data Analysis", IEEE ACM Transactions on Computational Biology and Bioinformatics, 4(3), p365-381, Jul), (2007.
[https://doi.org/10.1109/tcbb.2007.70224]

-
P. A. Mundra, and J. C. Rajapakse, "SVM-RFE With MRMR Filter for Gene Selection", IEEE Transactions on Nanobioscience, 9(1), p31-37, Oct), (2010.
[https://doi.org/10.1109/tnb.2009.2035284]

- C. Kim, "Feature Selection of SVM-RFE Combined with a TD Reinforcement Learning", Journal of KIIT. Vol. 16, 10, p21-26, Oct), (2018.
Feb., 2006 : Ph.D., Computer Science, Korea University
Nov., 2005. ~ Dec., 2008. : Senior Project Researcher, KISTI
Mar., 2009. ~ Feb., 2018. : Adjunct Professor, Dept. of Computer Science, Kyonggi University
Mar. 2018. ~ present : Assistant Professor, Div. of General Studies, Kyonggi University
Reserach Interest : Big-Data, Machine Learning, Deep Learning, Reinforcement Learning, Development of AI game